Our lab uses statistical and computational approaches to study questions in human genetics and evolutionary biology. A central aim in the lab is to understand the impact of evolutionary history on genetic variation and to apply this knowledge to learn about human history and biology. To this end, we use genetic data from ancient specimens and present-day individuals to study: (1) how different evolutionary processes such as mutation rate evolve across individuals, (2) when key events (such as introgression and adaptations) occurred in human history, and (3) how we can leverage these patterns to identify genetic variants related to human adaptation and disease. Some ongoing projects in the lab are described below.
Evolution of mutation rate across species
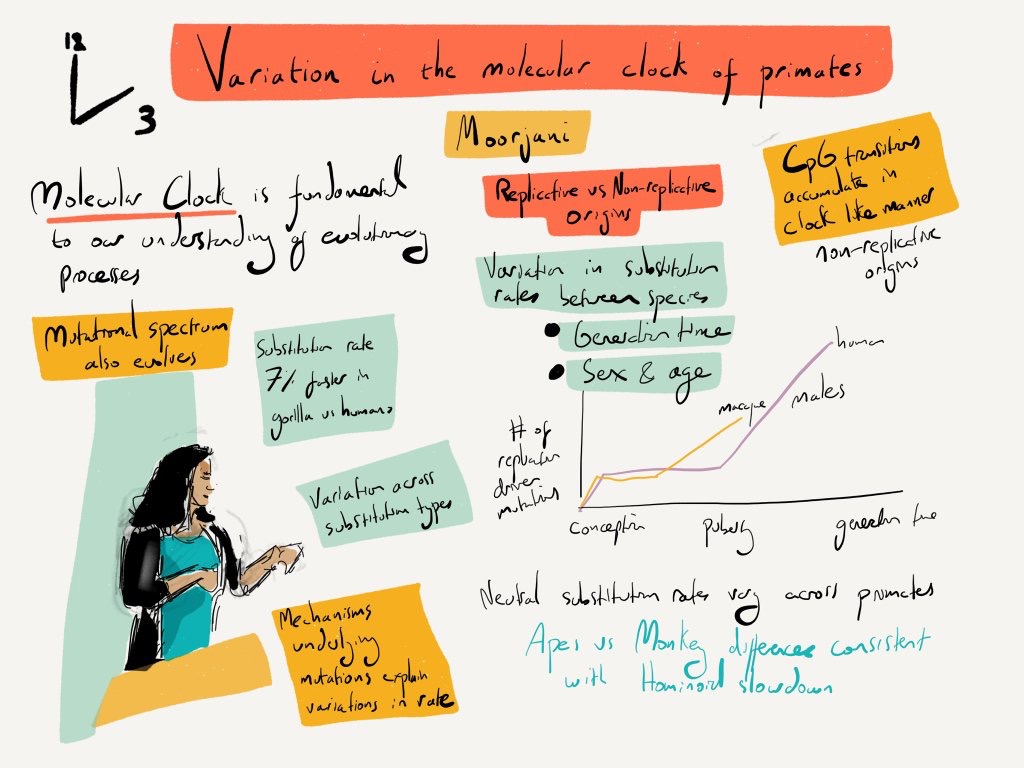
Image: Alex Cagan, CSHL 2016
Germline mutations are the ultimate source of genetic differences among individuals and across species; they provide the raw material for selection to act on, as well as play a role in many diseases. As mutations occur steadily over time, they provide a record of the time elapsed and hence a “molecular clock” for dating evolutionary events. However, despite strong constraints on the replication machinery, recent studies have shown that the mutation rate as well as the mutation spectra evolves rapidly across closely related species and also varies among humans. Thus, to investigate the causes of interspecies variation in mutation rate and to build robust models of evolution, we are interested in estimating direct pedigree-based mutation rates in humans and other primates. This will allow us to learn about the determinants of mutation rate and the mechanisms impacting its evolution across species.
Representative lab publications on this topic include:
- Gao Z+, Zhang Y, Cramer N, Przeworski M, Moorjani P+ (2022). Limited role of generation time changes in driving the evolution of the mutation spectrum in humans. eLife 12, e81188. DOI: https://doi.org/10.7554/eLife.81188.
- , , , , , A comparison of humans and baboons suggests germline mutation rates do not track cell divisions. PLoS biology. 18, e3000838.
- Chintalapati M and Moorjani P+. (2020). Evolution of the mutation rate across primates. Current Opinion in Genetics & Development. 62, 58-64.
- Moorjani P, Gao Z, and Przeworski M. (2016). Human germline mutation and the erratic evolutionary clock. PLoS Biol 14(10): e2000744. doi:10.1371/journal.pbio.2000744.
- Moorjani P , Amorim CE, Arndt P, and Przeworski M. (2016). Variation in the molecular clock in primates. Proc Natl Acad Sci USA. Sep 6. pii: 201600374.
Reconstructing population history using genetic data
A major focus of the research in the lab is to use DNA sequences from ancient specimens and present-day individuals to characterize population relationships and date evolutionary events. To this end, we are interested in: (a) developing new computational methods to reconstruct past evolutionary events, and (b) analyzing large-scale genomic datasets to uncover previously uncharacterized events. Previous work in this area has included characterizing population history in contemporary human populations, dating archaic admixture between humans and Neanderthals and estimating the divergence time of humans and apes. These, and other studies, highlight how genetic studies can complement historical and paleontological evidence to learn about human history over deep timescales.

Image: Manjusha Chintalapati
Representative lab publications on this topic include:
- Iasi LM*,†, Chintalapati M*,†, Skov L , Mesa AB , Hajdinjak M, Peter BM*,†, Moorjani P*,†. Neandertal ancestry through time: Insights from genomes of ancient and present-day humans. Science 386.6727 (2024): eadq3010.
- Kerdoncuff E*,†, Skov L*,†, Patterson N, Banerjee J, Khobragade P, Chakrabarti SS, Chakrawarty A, Chatterjee P, Dhar M, Gupta M, John JP, Koul PA, Lehl SS, Mohanty RR, Padmaja M, Perianayagam A 12, Rajguru C, Sankhe L, Talukdar A, Varghese M, Yadati SR, Zhao W, Leung YY, Schellenberg GD, Wang YZ, Smith JA, Dey S, Ganna A , Dey AB†, Kardia SLR†, Lee J†, Moorjani P†. 50,000 years of Evolutionary History of India: Impact on Health and Disease Variation. Cell 188 (2025):13, 3389-3404.e6.
- Chintalapati M+, Patterson N+, Moorjani P+. (2022) Reconstructing the spatiotemporal patterns of admixture during the European Holocene using a novel genomic dating method. eLife. DOI: https://doi.org/10.7554/eLife.77625.
- Tournebize R+, Chu G and Moorjani P+. (2022). Reconstructing the history of founder events using genome-wide patterns of allele sharing across individuals. PLoS Genetics. https://doi.org/10.1371/journal.pgen.1010243.
- Moorjani P, Sankararaman S, Fu Q, Przeworski M, Patterson N, Reich D. (2016). A genetic method for dating ancient genomes provides a direct estimate of human generation interval in the last 45,000 years. Proc Natl Acad Sci USA. May 17;113(20):5652-7. doi: 10.1073/pnas.1514696113.
Leveraging evolutionary history for identifying disease and adaptive variants
Characterizing population structure not only sheds light on population history, but also 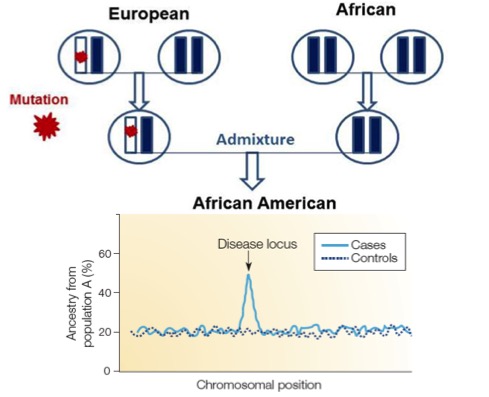 can be a very useful tool for uncovering disease genes. As disease prevalence often vary by ancestry, in populations of mixed ancestry such as African Americans and Latinos, we find that at the disease locus, cases have enhanced ancestry from the ancestral group with the higher disease risk. This approach called Admixture Mapping has been a powerful tool for identifying disease associations in recently admixed groups. We are interested in extending this approach to more complex admixed populations like South Asians, where the admixture is older and involves populations that no longer exist in unmixed form. In previous work, we showed that most Indian groups descend from a mixture of two genetically divergent populations: Ancestral North Indians (ANI) related to Central Asians, Middle Easterners, Caucasians, and Europeans; and Ancestral South Indians (ASI) not closely related to groups outside the subcontinent. This mixture occurred 1,900-4,200 years ago and was followed by a demographic shift towards endogamy, leading to extreme founder events in India. Both the history of founder events and admixture in India provide a unique opportunity for disease mapping as they predict the presence of rare, population specific disease variants that are not seen outside the subcontinent.
can be a very useful tool for uncovering disease genes. As disease prevalence often vary by ancestry, in populations of mixed ancestry such as African Americans and Latinos, we find that at the disease locus, cases have enhanced ancestry from the ancestral group with the higher disease risk. This approach called Admixture Mapping has been a powerful tool for identifying disease associations in recently admixed groups. We are interested in extending this approach to more complex admixed populations like South Asians, where the admixture is older and involves populations that no longer exist in unmixed form. In previous work, we showed that most Indian groups descend from a mixture of two genetically divergent populations: Ancestral North Indians (ANI) related to Central Asians, Middle Easterners, Caucasians, and Europeans; and Ancestral South Indians (ASI) not closely related to groups outside the subcontinent. This mixture occurred 1,900-4,200 years ago and was followed by a demographic shift towards endogamy, leading to extreme founder events in India. Both the history of founder events and admixture in India provide a unique opportunity for disease mapping as they predict the presence of rare, population specific disease variants that are not seen outside the subcontinent.
Representative lab publications on this topic include:
- Kerdoncuff E*,†, Skov L*,†, Patterson N, Banerjee J, Khobragade P, Chakrabarti SS, Chakrawarty A, Chatterjee P, Dhar M, Gupta M, John JP, Koul PA, Lehl SS, Mohanty RR, Padmaja M, Perianayagam A 12, Rajguru C, Sankhe L, Talukdar A, Varghese M, Yadati SR, Zhao W, Leung YY, Schellenberg GD, Wang YZ, Smith JA, Dey S, Ganna A , Dey AB†, Kardia SLR†, Lee J†, Moorjani P†. 50,000 years of Evolutionary History of India: Impact on Health and Disease Variation. Cell 188 (2025):13, 3389-3404.e6.
- , , , , , , , , Effect of APOE e4 and its modification by sociodemographic characteristics on cognitive measures in South Asians from LASI-DAD. Alzheimer’s Disease & Dementia.
- Moorjani P, Thangaraj K, Patterson N, Lipson M, Loh PR, Govindaraj P, Berger B, Reich D, Singh L. Genetic evidence for recent population mixture in India. (2013). American Journal of Human Genetics. Sep 5;93(3):422-38. doi: 10.1016/j.ajhg.2013.07.006.
- , , , , ,, , , , , , , , , , , K. (2017). The promise of disease gene discovery in South Asia. Nature Genetics, doi:10.1038/ng.3917.